Levelset optimization#
In this notebook we’ll carry out optimization of a the ceviche power splitter challenge using a levelset parameterization. With the levelset parameterization, the boundaries of features will be free to move, but the topology should remain unchanged.
Begin by creating the ceviche_power_splitter challenge and obtaining initial parameters.
from invrs_gym import challenges
challenge = challenges.ceviche_power_splitter()
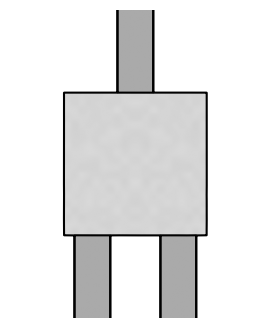
The default initial parameters are randomly generated, as is typical in a topology optimization setting. Let’s visualize these:
import jax
import matplotlib.pyplot as plt
from skimage import measure
default_params = challenge.component.init(jax.random.PRNGKey(0))
def plot_params(ax, params, contour_levels=(0.5,)):
density = challenge.component.ceviche_model.density(params.array)
im = ax.imshow(1 - density, cmap="gray")
im.set_clim(-2, 1)
for level in contour_levels:
for c in measure.find_contours(density, level=level):
plt.plot(c[:, 1], c[:, 0], 'k')
_ = ax.axis(False)
plt.figure(figsize=(6, 4))
plot_params(ax=plt.subplot(111), params=default_params, contour_levels=(0.4, 0.6))
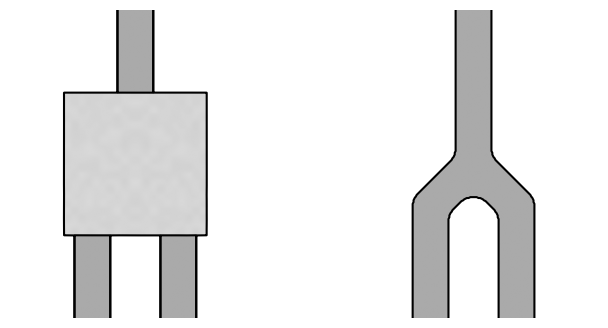
In this notebook, we’ll construct our own initial parameters, which will be more suited to levelset optimization. Define an auto_route function for this purpose, and then visualize the generated structure.
import dataclasses
import numpy as onp
from skimage import morphology
from imageruler import imageruler
from totypes import types
def auto_route(density: types.Density2DArray) -> types.Density2DArray:
"""Automatically routes waveguides in a design region."""
assert density.ndim == 2
array = onp.ones_like(density.array, dtype=bool)
if density.fixed_void is not None:
array[density.fixed_void] = False
d = max(array.shape)
array = onp.pad(array, pad_width=((d, d), (d, d)), mode="edge")
array = morphology.skeletonize(array, method="lee")
array = array.astype(bool)
length_scale = border_structure_length_scale(density)
kernel = imageruler.kernel_for_length_scale(length_scale)
array = morphology.binary_dilation(array[:, ::-1], kernel)[:, ::-1]
array = morphology.binary_closing(array[:, ::-1], kernel)[:, ::-1]
array = morphology.binary_opening(array[:, ::-1], kernel)[:, ::-1]
array = array[d:-d, d:-d]
if density.fixed_solid is not None:
array[density.fixed_solid] = True
if density.fixed_void is not None:
array[density.fixed_void] = False
array = onp.where(array == 0, density.lower_bound, density.upper_bound)
density = dataclasses.replace(density, array=array)
return types.symmetrize_density(density)
def border_structure_length_scale(density: types.Density2DArray) -> int:
"""Finds the length scale of structures at the border of a density array."""
array = onp.array(density.array)
if density.fixed_solid is not None:
array[density.fixed_solid] = density.upper_bound
if density.fixed_void is not None:
array[density.fixed_void] = density.lower_bound
array = array > (density.lower_bound + density.upper_bound) / 2
borders = (array[:, 0], array[:, -1], array[0, :], array[-1, :])
segment_length_scales = [
min(imageruler.minimum_length_scale_1d(border_segment, periodic=False))
for border_segment in borders
]
return min(segment_length_scales)
params = auto_route(default_params)
plt.figure(figsize=(8, 4))
plot_params(ax=plt.subplot(121), params=default_params, contour_levels=(0.4, 0.6))
plot_params(ax=plt.subplot(122), params=params)
To carry out levelset optimization, we can use the levelset_wrapped_optax optimizer from the invrs-opt library. This optimizer uses a user-specified optax optimizer to minimize an objective, where the optimization variables include density arrays parameterized via a levelset function.
import invrs_opt
import optax
opt = invrs_opt.levelset_wrapped_optax(optax.adam(0.1), penalty=1.0)
state = opt.init(params)
@jax.jit
def step_fn(state):
def loss_fn(params):
response, aux = challenge.component.response(params)
eval_metric = challenge.eval_metric(response)
return challenge.loss(response), (response, aux, eval_metric)
params = opt.params(state)
(loss, (response, aux, eval_metric)), grad = jax.value_and_grad(loss_fn, has_aux=True)(params)
state = opt.update(grad=grad, value=loss, params=params, state=state)
return state, (params, response, loss, aux, eval_metric)
for i in range(50):
state, (params, response, loss, aux, eval_metric) = step_fn(state)
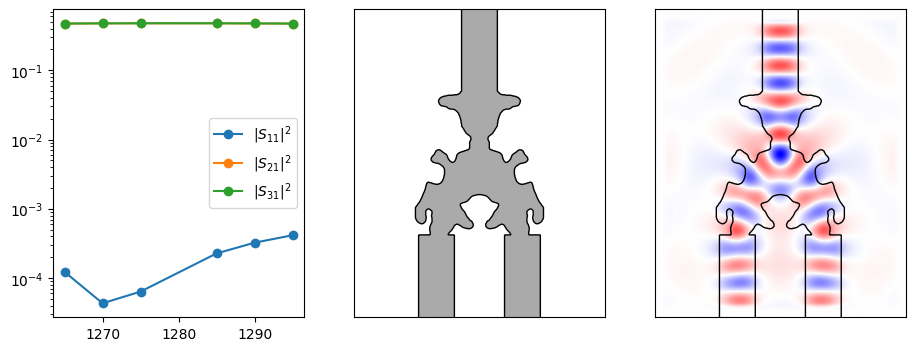
After running the optimization, we can now visualize the final structure and its performance.
def plot_ceviche_component(component, params, response, aux):
plt.figure(figsize=(11, 4))
ax = plt.subplot(131)
for i in range(response.s_parameters.shape[-1]):
ax.semilogy(
response.wavelengths_nm,
onp.abs(response.s_parameters[:, 0, i]) ** 2,
"o-",
label="$|S_{" + f"{i + 1}1" + "}|^2$",
)
ax.legend()
# Get the full structure, including waveguides extending away from the deisgn.
density = component.ceviche_model.density(params.array)
contours = measure.find_contours(density)
ax = plt.subplot(132)
im = ax.imshow(1 - density, cmap="gray")
im.set_clim([-2, 1])
for c in contours:
plt.plot(c[:, 1], c[:, 0], "k", lw=1)
ax.set_xticks([])
ax.set_yticks([])
ax = plt.subplot(133)
fields = onp.real(aux["fields"][2, 0, :, :])
im = ax.imshow(fields, cmap="bwr")
im.set_clim([-onp.amax(onp.abs(fields)), onp.amax(onp.abs(fields))])
for c in contours:
plt.plot(c[:, 1], c[:, 0], "k", lw=1)
ax.set_xticks([])
ax.set_yticks([])
plot_ceviche_component(challenge.component, params, response, aux)